ADVERTISEMENT
Assessing The Impact Of Biofilm And The Microbiome In Diagnosing And Treating DFUs
 The incidence of diabetes mellitus has increased at a dramatic rate, affecting a reported 29.1 million Americans.1 More significantly, an estimated 50 percent of Americans have diabetes or pre-diabetes.2 An estimated 15 percent of individuals with diabetes will develop a lower extremity ulceration in their lifetime with many of these ulcerations progressing into chronic wounds, which may become infected, require multiple hospitalizations and, more alarmingly, result in amputations.3
The incidence of diabetes mellitus has increased at a dramatic rate, affecting a reported 29.1 million Americans.1 More significantly, an estimated 50 percent of Americans have diabetes or pre-diabetes.2 An estimated 15 percent of individuals with diabetes will develop a lower extremity ulceration in their lifetime with many of these ulcerations progressing into chronic wounds, which may become infected, require multiple hospitalizations and, more alarmingly, result in amputations.3
The initial evaluation and treatment of a diabetic foot ulcer involves debridement of all necrotic and non-viable tissue; wound exploration to determine the depth and extent of tissue involved; and diagnosis of infection, if present. Traditionally, the determination of wound infection includes assessment of any clinical signs of infection such as erythema, edema, warmth and pain. It also includes laboratory tests and collecting culture swabs of the wound with deep cultures preferred. Plain radiographs and advanced imaging may be necessary to evaluate for osteomyelitis or fluid collections.
Proper treatment of infected wounds involves the drainage of all purulent collections if present and appropriately directed antibiotic therapy. Physicians can administer antibiotic therapy through enteral, parenteral and topical routes, or any combination. In the infected diabetic foot ulcer, initial treatment with broad spectrum coverage of possible pathogens is advised until culture data can provide information to target the specific organisms responsible for the infection.
Comparing Wound Culturing With DNA Sequencing
Rapid and accurate identification of the pathogens responsible for wound infections is paramount for diagnosis and treatment. Conventional identification of bacteria uses phenotypic tests relying on growth on culture medium, leading to major limitations in the proper identification of bacteria. These limitations include the growth of organisms that grow abundantly in customary culture media and not necessarily the most relevant or clinically important pathogen. Authors widely acknowledge that reliance on standard culture techniques has led to a gross underestimation of the bacteria present in the chronic wound with these cultures detecting roughly 1 percent of the bacteria present.4
The advent of gene sequencing of 16S rDNA has revealed vastly more complex bacterial communities than those identified by traditional culture methods, particularly in chronic wounds.5 Since 2001, the use of gene sequencing has led to the identification of 215 novel bacterial species. Furthermore, the use of gene sequencing has allowed the identification of the normal microbial makeup of healthy human beings and led to a greater understanding of the important role normal bacterial flora hold in preventing illness and disease.6
In 2001, Lederberg coined the term microbiome to describe the “ecological community of commensal, symbiotic and pathogenic microorganisms that literally share our body space.”7 Historically, microbiologic studies have focused on disease causing organisms with little focus on organisms that routinely inhabit the human body. In 2005, the National Institutes of Health started an initiative called the Human Microbiome Project, consisting of multiple projects with the goal of identifying and characterizing the microorganisms found in the normal human microbiome.
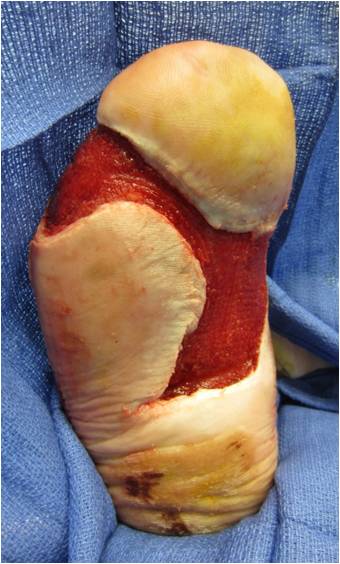 With the increasing availability of DNA sequencing technology, researchers have investigated the microbiome of normal skin and that of chronic wounds such as diabetic foot ulcers to determine if the microbiome presents a barrier to wound healing. Hoiby and colleagues have suggested that chronic wounds are, in fact, chronic wound infections secondary to the presence and activities of the biofilm.8 The importance of biofilm is underscored by its presence in over 90 percent of chronic wounds.9
With the increasing availability of DNA sequencing technology, researchers have investigated the microbiome of normal skin and that of chronic wounds such as diabetic foot ulcers to determine if the microbiome presents a barrier to wound healing. Hoiby and colleagues have suggested that chronic wounds are, in fact, chronic wound infections secondary to the presence and activities of the biofilm.8 The importance of biofilm is underscored by its presence in over 90 percent of chronic wounds.9
Wound biofilms are a polymicrobial group of cells attached to the surface of the wound, encasing themselves in an extracellular polymeric substance and producing inflammatory cytokines and matrix metalloproteinases (MMP) that impede the normal secretion of growth factors and chemoattractants. While antibiotic resistance is not an issue with this polymicrobial group of cells, variations in phenotype have resulted in a recalcitrant wound bed with diminished metabolic activity.10 Physicians can effectively combat the biofilm with sharp debridement, biofilm-targeted therapies and antibiotics.
However, there is increasing concern that the use of antibiotics to address the biofilm is instead compromising the natural microbiome and contributing to the observed bacterial antibiotic resistance.11 Furthermore, the extent to which microorganisms impair wound healing remains an ongoing area of debate in the management of chronic wounds. Recent studies evaluating the diabetic foot ulcer microbiome could provide a deeper understanding of the role of the microbiome in chronic wounds, potentially guiding new strategies to effectively control the growth of polymicrobial biofilms and improve healing.12-15
Certainly, we have underestimated the polymicrobial nature of chronic wounds with the use of culture-based wound sampling. Therefore, it is possible that wound care management, when based on incomplete diagnostics, may lead to suboptimal treatment. Technology has now made it possible to improve the capacity of microbial identification as well as detection of genetic elements to evaluate virulence and resistance patterns to help define and guide treatment.
Gene sequencing has provided several key findings regarding the microbiological environment of diabetic foot ulcers. First, the sensitivity of traditional wound culturing techniques in comparison to DNA sequencing is relatively low with the complexity of the bacterial population underrepresented from culture technique with cultures frequently missing anaerobic isolates.12 Additionally, in one study, increased ulcer duration positively correlated with wound flora diversity while increased ulcer depth demonstrated increased anaerobes.13
Can Polymerase Chain Reaction Testing Be A Game Changer With Antibiotic Management?
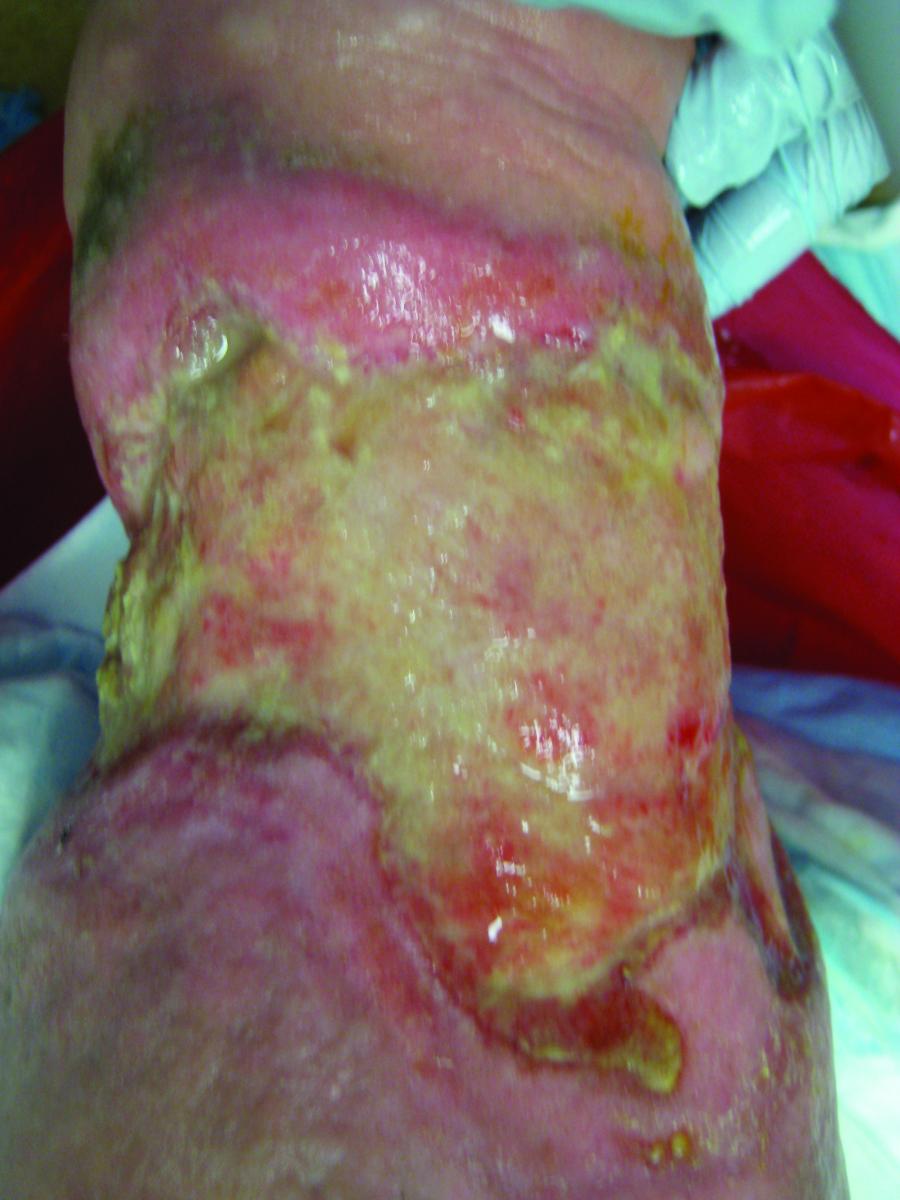 Armed with knowledge of the increased wound flora diversity, one may initiate antibiotic treatment based on specific genetic virulence and resistance patterns to provide improved treatment. Topical antibiotics based upon polymerase chain reaction (PCR) testing have become a popular treatment choice for the rapid detection of biofilm and antibiotic-resistant isolates.14 Obtain a swab of the ulceration and send it to a lab for DNA-based PCR testing. The resulting information helps technicians formulate a specific topical antibiotic based on the exact pathogen and virulent patterns present. This process is faster and more efficient than traditional culture swabs that require waiting for the pathogens to grow and then performing additional testing for sensitivities.
Armed with knowledge of the increased wound flora diversity, one may initiate antibiotic treatment based on specific genetic virulence and resistance patterns to provide improved treatment. Topical antibiotics based upon polymerase chain reaction (PCR) testing have become a popular treatment choice for the rapid detection of biofilm and antibiotic-resistant isolates.14 Obtain a swab of the ulceration and send it to a lab for DNA-based PCR testing. The resulting information helps technicians formulate a specific topical antibiotic based on the exact pathogen and virulent patterns present. This process is faster and more efficient than traditional culture swabs that require waiting for the pathogens to grow and then performing additional testing for sensitivities.
It is well recognized that treatment of any underlying infection is essential for proper management of a chronic diabetic foot ulcer. Should one use prophylactic antibiotics to treat a chronic ulcer when there are concerns about a biofilm presence impeding wound healing?
One recent study evaluated the effect of oral antibiotic therapy on wound healing in mice.15 Researchers compared the wound healing of full-thickness wounds between mice treated with antibiotics for one week and control mice. Surprisingly, the investigators found that antibiotic fed mice had significantly slower wound healing and, more importantly, antibiotic treatment decreased the bacterial density and altered the bacterial composition in the epithelialization of the wounds after injury. While the results of this study do not discourage the use of appropriate antibiotics for the treatment of wound infection, they do give us pause to consider the possible negative effects associated with microbiome alteration and delayed wound healing.
In Conclusion
As the advent of DNA technology has advanced our understanding of the microbiome and its beneficial role in preventing disease, it has also prompted us to question our traditional paradigms of treatment for the chronic diabetic foot ulcer.
Dr. Dinh is an Assistant Professor in Surgery at Harvard Medical School. She is also affiliated with the Beth Israel Deaconess Medical Center in Boston. Dr. Dinh is a Fellow and a member of the Board of Directors of the American College of Foot and Ankle Surgeons.
Dr. Lundborg is a first-year resident within the Podiatric Surgical Residency Program at the Beth Israel Deaconess Medical Center.
References
- American Diabetes Association. Economic costs of diabetes in the U.S in 2012. Diabetes Care. 2013; 36(4):1033–1046.
- Menke A, Casagrande S, Geiss L, Cowie C. Prevalence of and trends in diabetes among adults in the United States, 1988-2012. J Am Med Assoc. 2015; 314(10):1021–1029.
- Redel H, Gao Z, Li H, Alekseyenko A, et al. Quantification and composition of cutaneous microbiota in diabetic and nondiabetic men. J Infect Dis. 2013; 207(7):1105–1114.
- Martin JM, Zenilman JM, Lazarus GS. Molecular microbiology: new dimensions for cutaneous biology and wound healing. J Investig Dermatol. 2010;130(1):38–48.
- Dowd SE, Sun Y, Secor PR, et al. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008; 8:43.
- Dowd SE, Wolcott RD, Sun Y, McKeehan T, Smith E, Rhoads D. Polymicrobial nature of chronic diabetic foot ulcer biofilm infections determined using bacterial tag encoded FLX amplicon pyrosequencing (bTEFAP). PLoS One. 2008; 3(10):e3326.
- Lederberg J, McCray AT. ‘Ome sweet ‘omics - a genealogical treasury of words. Scientist. 2001;15(7):8–8.
- Hoiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015; 21(Suppl1):S1–25.
- Attinger C, Wolcott R. Clinically addressing biofilm in chronic wounds. Wound Healing Society. Adv Wound Care. 2012; 1(3):127–32.
- Lewis K. Riddle of biofilm resistance. Antimicrob Agents Chemother. 2001;45:999–1007.
- Howell-Jones RS, Price PE, Howard AJ, Thomas DW. Antibiotic prescribing for chronic skin wounds in primary care. Wound Repair Regen. 2006; 14(4):387–93.
- Gardner SE, Hillis SL, Heilmann K, Segre JA, Grice EA. The neuropathic diabetic foot ulcer microbiome is associated with clinical factors. Diabetes. 2013;62(3):923–930.
- Salisbury S, Sabatini L, Spiegel C. Identification of methicillin-resistant staphylococci by multiplex polymerase chain reaction assay. Am J Clin Pathol. 1997; 107(3):368–73.
- Zhang M, Jiang Z, Li D, Jiang D, Wu Y, Ren H, Peng H, Lai Y. Oral antibiotic treatment induces skin microbiota dysbiosis and influences wound healing. Microb Ecol. 2015; 69(2):415-21.











