Prevalence of Silver Resistance in Bacteria Isolated from Diabetic Foot Ulcers and Efficacy of Silver-Containing Wound Dressings
Silver is an effective, broad-spectrum antimicrobial agent commonly incorporated into topical creams (eg, silver sulfadiazine)1-11 and wound dressings to manage wound infection. Despite the medical benefits of using ionic silver to manage infections, concern has been raised regarding the potential for development of bacterial resistance and an association with cross-resistance to antibiotics has been implied. Cross-resistance is of concern because plasmids encoding silver-resistant genes also may encode for antibiotic resistance.12,13
Silver-resistant organisms have been reported in clinical5,14-18 and environmental 19-22 samples. The genetic basis for silver resistance was first reported by McHugh et al13 underscoring that silver resistance was plasmid-encoded. This conclusion has been confirmed by others.16,22-25
The physiological, biochemical, genetic, and structural studies of the silver-resistant determinant plasmid pMG101 established the molecular basis of silver resistance.12,26-29 Plasmid pMG101 is a 182 kb, transferable plasmid26 encoding resistance to silver (nine Open Reading frames [ORFs] in three transcriptional units), mercury, tellurite, ampicillin, chloramphenicol, tetracycline, streptomycin, and sulphonamide.13,26 It confers resistance in bacteria at silver concentrations six or more times the concentration of what a sensitive Escherichia coli can tolerate.30 Functions assigned to the genes are based on homologous systems encoding resistances to other metals. The silver-resistance system encodes two silver efflux pumps (one an ATPase and the other chemiosmotic) and two periplasmic Ag+-binding proteins.31 The genes associated with silver resistance provided the basis of the current investigation to determine the prevalence of silver resistance genes in clinical wound isolates.
The goals of this study were 1) to investigate the presence and prevalence of silver resistance and silver-resistant genes in clinical wound isolates taken from diabetic foot ulcers and 2) to examine the effects of silver-containing dressings on resistant bacterial viability.
Methods
Organism collection and identification. The micro-organisms used in this study were routinely isolated from the chronic foot wounds of patients attending the Diabetic Foot Clinic at Tameside General Hospital, UK who had diabetic foot ulcers exhibiting signs and symptoms of clinical infection, as well as ulcers clinically free from infection. All wounds were swabbed using calcium alginate swabs. All microorganisms were cultured and identified at Medi-Lab Ltd, UK using standard protocols. The isolates obtained for this study were coagulase negative Staphylococcus aureus (CNS – 17 strains), Staphylococcus aureus (24 strains), Enterobacter spp. (three strains), Enterobacter cloacae (four strains), Escherichia coli (six strains), additional coliforms (two strains), Pseudomonas spp. (six strains), Pseudomonas aeruginosa (nine strains), Enterococcus faecalis (five strains), Alcaligenes faecalis (two strains), diphtheroids (10 strains), Citrobacter spp. (one strain), Proteus mirabilis (two strains), Serratia marcescens (one strain), Acinetobacter calcoaceticus (one strain), Providencia rettgeri (two strain), Streptococcus haemolyticus (one strain), Morganella morganii (two strains), Peptostreptococcus prevotti (three strains), Peptostreptococcus tetradius (one strain), Peptostreptococcus micros (one strain), and nine other unclassified bacteria. All bacteria were screened for resistance to silver.
In vitro studies.
Media, chemicals, primers, and culture methods. Unless otherwise stated, all clinical wound isolates were maintained on Mueller Hinton agar (MHA, Oxoid, UK). Recombinant positive controls were maintained on Luria Bertani media (Sigma Aldrich, Mo) that selected for 100 µg/mL ampicillin. Oligonucleotide primers used in this study were derived from those used by Gupta et al13 in an in vitro study of bacterial survival in response to silver, purchased from MWG-BIOTECH (Ebersberg, Germany), with the following composition: silE gene: Reverse GGCCAGACTGACCGTTATT and forward GTACTC CCCCGGACATCACTAATT; silS gene reverse GTTTGCTGCATGACAGGCTAA AGACATC and forward GGAGATCCCGGATGCATAGCAA; silP gene Reverse CGGGCAGACCAGCAATAACAGATA and forward CATGACATATCCTGAAGA CAGAAAATGC. (The letters represent DNA bases in a custom-designed oligonucleotide sequence complementary to the sequence of genes examined.) Polymerase chain reaction (PCR) was performed using Reddymix PCR mastermix (Abgene, Epsom, UK) and contained primers at a final concentration of 24 to 34 pmol together with 3 µl of DNA template (50 µl final volume).
Controls. Positive control strains obtained from Gupta et al26 included E. coli strain J53 containing plasmid pMG10117 and cloned fragments from that plasmid. Plasmid pKM1 contains an ampicillin resistance marker and the silS and silE genes were transferred into E. coli strain DH5α. DH5α(pKM1) shows partial resistance to Ag+.11 For the silP gene control, plasmid pKM326 with an ampicillin marker was used. A plasmid preparation of the E. coli strains DH5α and DH5α(pKM3)26 provided an additional positive PCR control. E. coli J53 without a plasmid was the negative control for all three genes.
Polymerase chain reaction (PCR). A colony from each streaked clinical wound isolate grown on MHA was diluted in 10 µL of sterile water and heated at 95° C for 5 minutes, vortexed, and then centrifuged briefly. The supernatant fluid after centrifugation was used as the PCR template. A PCR reaction mixture of the following composition was prepared in a 50 µL microfuge tube: 41 µL Reddymix PCR mastermix, 3 µL of forward primer solution, 3 µL reverse primer solution, 3 µL of template preparation, and a drop of mineral oil (Sigma Aldrich, Mo). The PCR was run using a Perkin Elmer DNA Thermal Cycler 480 (US instrument division, Norwalk, Conn) programmed at an initial 95° C for 2 minutes, then 40 cycles of 95° C for 1 minute, 55° C for 1 minute, and 72° C for 3 minutes, followed finally by 72° C for 5 minutes and storage at 4° C. The PCR program consisted of standard steps at temperatures and durations relevant to the size of PCR products produced.13
Agarose gel electrophoresis. The gel wells were filled with 12 µl of PCR products from the wound isolates and controls with a 1 kb plus Ready Load DNA Marker “ladder” (Invitrogen, Calif) providing size standards. Electrophoresis was run at 65 V for 90 minutes; the gel then was photographed with a Gene Genius Bioimaging System (Syngene, Synoptics Ltd Cambridge, UK). All experiments were carried out in duplicate.
Sequencing. The PCR products were excised from the agarose gel and purified using the Qiagen Q1A quick PCR purification kit, visualized on a 1.5% agarose gel, diluted to 10 ng/µL with water, and sequenced by the Advanced Biotechnology Centre, Imperial College London, using ABI 3100 16 capillary genetic analyzers. Sequence results were analyzed using the NCBI BlastN program.
Test materials. A silver-containing Hydrofiber® (SCH, ConvaTec Wound Therapeutics, Deeside, UK) dressing (100% sodium carboxymethylcellulose with 1.2% w/v ionic silver), a non-silver-containing Hydrofiber® (NSH) dressing, and a nanocrystalline silver-containing alginate dressing (NCS, Smith & Nephew, UK) were assessed. Two different silver-containing dressings were used in this in vitro study in order to examine the effects silver on the survival of bacteria shown to contain resistance genes.
Confocal microscopy. All strains shown to be sil positive, together with a non-silver-resistant P. aeruginosa and S. aureus, were investigated using this method. A single bacterial colony was taken from a 24-hour agar plate and suspended in Maximal Recovery Diluent (MRD, Oxoid, UK) to a concentration corresponding to a McFarland Standard of 3.0. Molecular Probes LIVE/DEAD Baclight™ stain (Invitrogen, Paisley, UK) was made following manufacturers instructions in 5 mL of distilled water and mixed thoroughly. This solution was stored in the dark at room temperature until required. Subsequently, 500 µL of Baclight™ stain was gently mixed with 500 µL of suspended bacteria and incubated in a dark at room temperature for at least 10 minutes and 1-cm2 samples of a NSH, SCH, and a NCS were cut aseptically and placed on coverslips suspended over a 4-cm2 hole cut into the base of a Petri dish. Using sterile forceps, the dressings were teased apart to expose fibers to aid visualization. To each of the dressings, 150 µL of Baclight™-stained bacteria and up to 50 µL of additional MRD to saturate the dressing were added. The dish was covered and kept in the dark until ready for confocal imaging.
Samples were imaged using a Leica TCS SP2 inverted rapid-scanning confocal laser microscope set to image wavelengths between 500 to 530 nm (Syto9 stains viable cells) and 610 to 640 nm (presidium's iodide [PI] stains dead cells) on two separate PMT channels.32 Images were captured at 3, 24, and 48 hours after dressings were inoculated.
Results
The 112 bacteria isolated from diabetic foot ulcers were screened for the presence of the silS, silE, and silP genes. Of the total number of isolates, 1.8% was found to contain silver resistance genes. Two strains were found to be positive for these genes – both E. cloacae (strains assigned as Z953109 and Z959947). The agarose gel electrophoresis of PCR products for silS, silE, and silP genes from these two E. cloacae isolates are shown in Figure 1. Both E. cloacae strains contained the silver-resistance genes silE, silS, and silP. A third strain of E. cloacae (CN – kindly donated by Alan Lansdown,  Imperial College, London)33 that previously had been shown to grow in the presence of >50 µg/mL silver nitrate) also was screened in this study as a positive control. The CN isolate was positive for silE, silS, and silP genes as shown in Figure 1. The sequence from the PCR products for the silE, silS, and silP genes shared the same homology (96%) with the sil genes from pMG101 (NCIB Accession number AG067954 - Gupta et al13) and 100% homology with E. cloacae Ag703 (NCIB 679159).
Imperial College, London)33 that previously had been shown to grow in the presence of >50 µg/mL silver nitrate) also was screened in this study as a positive control. The CN isolate was positive for silE, silS, and silP genes as shown in Figure 1. The sequence from the PCR products for the silE, silS, and silP genes shared the same homology (96%) with the sil genes from pMG101 (NCIB Accession number AG067954 - Gupta et al13) and 100% homology with E. cloacae Ag703 (NCIB 679159).
Incubation of silver-resistant strains Z953109, Z959947, and CN with SCH and NCS revealed that more than 95% of all bacteria were completely killed after 48 hours (see Figure 2e,f). Images shown are of strain Z959947 and are representative of cell death seen in all genetically resistant strains. Evidence of bacterial cell death (red PI-stained cells) appeared 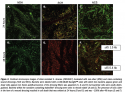 within 24 hours incubation with the silver-containing dressings (see Figure 2b,c). Examination of resistant strains with NSH showed that in the absence of ionic silver, bacteria were viable for the course of the study (see Figure 2a,d). Control strains (P. aeruginosa and S. aureus [data not shown]) were killed by the silver-containing dressings over a similar time period as resistant strains (see Figure 3a-f). Confocal microscopy allowed the samples to be viewed three dimensionally and showed that cell viability in the presence/absence of ionic silver was uniform throughout the dressings.
within 24 hours incubation with the silver-containing dressings (see Figure 2b,c). Examination of resistant strains with NSH showed that in the absence of ionic silver, bacteria were viable for the course of the study (see Figure 2a,d). Control strains (P. aeruginosa and S. aureus [data not shown]) were killed by the silver-containing dressings over a similar time period as resistant strains (see Figure 3a-f). Confocal microscopy allowed the samples to be viewed three dimensionally and showed that cell viability in the presence/absence of ionic silver was uniform throughout the dressings.
Discussion
Evidence of silver resistance in bacteria was first documented in burn wards.13-14,16 Most recently, evidence of silver-resistant genes in E. cloacae isolated from biofilms on teeth has been highlighted.23 To date, the prevalence of genetic 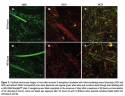 resistance to silver is not known, specifically with bacteria isolated from clinical settings where high levels of silver may be used. As silver is used widely in wound care, it is appropriate to determine the prevalence of silver resistance genes in wound pathogens.
resistance to silver is not known, specifically with bacteria isolated from clinical settings where high levels of silver may be used. As silver is used widely in wound care, it is appropriate to determine the prevalence of silver resistance genes in wound pathogens.
In this study, 112 clinical isolates obtained from patients with diabetic foot ulcers were screened by PCR for the presence of the silP, silE, and silS genes. Three E. cloacae (Z953109, Z959947 and CN) isolated from diabetic foot ulcers showed PCR products similar to that of pMG101 for all the three genes investigated.
Despite the fact that chronic wounds such as diabetic foot ulcers are often polymicrobial,34 it is interesting that in this study only strains of E. cloacae were sil positive. Bacteria considered to be notorious pathogens in wounds – eg, S. aureus and P. aeruginosa – were not associated with genetic resistance to silver, despite the significant number of these strains screened (nine P. aeruginosa and 24 S. aureus). In contrast, E. cloacae is not a primary pathogen in wounds, despite its frequent presence. Although a polymicrobial wound environment is likely to be conducive to the transfer of genetic resistance between species, no evidence of transfer of sil genes from E. cloacae was found in this study. Overall, the prevalence of silver resistance (sil positive) in bacteria isolated from diabetic foot ulcers was 1.6%, which is slightly higher than that previously observed in a burn wound that was measured by phenotypic resistance.35
Visualization of sil positive E. cloacae strains utilizing confocal microscopy demonstrated that despite the presence of genes associated with silver resistance, ionic silver, provided in the form of a silver-containing wound dressing (SCH and NCS), was able to eradicate the bacteria within 24 to 48 hours of incubation. Wear time for the dressings tested is usually 2 to 3 days, which is adequate time for the silver in the dressings to kill the sil gene-positive bacteria.
Conclusion
This study has shown that the prevalence of silver resistance genes in bacteria isolated from diabetic foot ulcers is low and appears to be confined to an enteric bacterium, E. cloacae, which is not known to be a primary wound pathogen. No evidence of silver resistance genes in wound pathogens such as S. aureus and P. aeruginosa was found in this study. Investigation of expression of these and other heavy metal-resistance genes should be carried out to elucidate the role each gene plays in conferring resistance.
This study also has shown that the low proportion of wound bacteria that possess silver resistance genes are killed when challenged with silver-containing wound dressings. Consequently, genetic resistance does not necessarily translate into phenotypic resistance.
Overall, this study has provided further evidence that the prevalence of genetic resistance to silver is low and that silver-containing wound dressings still can be effective in controlling such bacteria. It is the authors’ opinion that, while silver resistance in wound care should be continually monitored, the threat of widespread resistance is low and silver-containing dressings remain an extremely important tool in managing infected wounds and those at risk of infection.
Acknowledgment
The authors thank Simon Silver for technical assistance in compiling this manuscript.
1. Fox CL Jr, Rao TN, Azmeth R, Gandhi SS, Modak S. Comparative evaluation of zinc sulfadiazine and silver sulfadiazine in burn wound infection. J Burn Care Rehabil. 1990;11(2):112-117.
2. Fox CL Jr. Silver sulfadiazine–a new topical therapy for Pseudomonas in burns. Therapy of Pseudomonas infection in burns. Arch Surg. 1968;96(2):184-188.
3. George N, Faoagali J, Muller M. Silvazine (silver sulfadiazine and chlorhexidine) activity against 200 clinical isolates. Burns. 1997;23(6):493-495.
4. Hoffmann S. Silver sulfadiazine: an antibacterial agent for topical use in burns. A review of the literature. Scand J Plast Reconstr Surg. 1984;18(1):119-126.
5. Klasen HJ. A historical review of the use of silver in the treatment of burns. II. Renewed interest for silver. Burns. 2000;26(2):131-138.
6. Miller L, Hansbrough J, Slater H, et al. Sildimac: a new delivery system for silver sulfadiazine in the treatment of full-thickness burn injuries. J Burn Care Rehabil. 1990;11(1):35-41
7. Modak S, Fox P, Stanford J, Sampath L, Fox CL Jr. Silver sulfadiazine-impregnated biologic membranes as burn wound covers. J Burn Care Rehabil. 1986;7(5):422-425.
8. Modak SM, Sampath L, Fox C Jr. Combined topical use of silver sulfadiazine and antibiotics as a possible solution to bacterial resistance in burn wounds. J Burn Care Rehabil. 1988;9(4):359-363.
9. Monafo WW, Freedman B. Topical therapy for burns. Surg Clin North Am. 1987;67(1):133-145.
10. Monafo WW, West MA. Current treatment recommendations for topical burn therapy. Drugs. 1990;40(3):364-373.
11. Pruitt BA Jr, McManus AT, Kim SH, Goodwin CW. Burn wound infections: current status. World J Surg. 1998;22(2):135-145.
12. Gupta A, Phung LT, Taylor DE, Silver S. Diversity of silver resistance genes in IncH incompatibility group plasmids. Microbiology. 2001;147(Pt 12):3393-3402.
13. McHugh GL, Moellering RC, Hopkins CC, Swartz MN. Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin. Lancet. 1975;1(7901):235-240.
14. Annear DI, Mee BJ, Bailey M. Instability and linkage of silver resistance, lactose fermentation, and colony structure in Enterobacter cloacae from burn wounds. J Clin Pathol. 1976;29(5):441-443.
15. Carr HS, Rosenkranz HS. R factor in Enterobacter cloacae resistant to silver sulfadiazine. Chemotherapy. 1975;21(1):41-44.
16. Hendry AT, Stewart IO. Silver-resistant Enterobacteriaceae from hospital patients. Can J Microbiol. 1979;25(8):915-921.
17. Markowitz SM, Smith SM, Williams DS. Retrospective analysis of plasmid patterns in a study of burn unit outbreaks of infection due to Enterobacter cloacae. J Infect Dis. 1983;148(1):18-23.
18. Slots J, Feik D, Rams TE. Prevalence and antimicrobial susceptibility of Enterobacteriaceae, Pseudomonadaceae and Acinetobacter in human periodontitis. Oral Microbiol Immunol. 1990;5(3):149-154.
19. Belly RT, Kydd GC. Silver resistance in microorganisms. Develop Industrial Microbiol. 1982;23:567-577.
20. Choudhury P, Kumar R. Multidrug- and metal-resistant strains of Klebsiella pneumoniae isolated from Penaeus monodon of the coastal waters of deltaic Sundarban. Can J Microbiol. 1998;44(2):186-189.
21. Grewal JS, Tiwari RP. Resistance to metal ions and antibiotics in Escherichia coli isolated from foodstuffs. J Med Microbiol. 1990;32(4):223-226.
22. Haefeli C, Franklin C, Hardy K. Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine. J Bacteriol. 1984;158(1):389-392.
23. Davis IJ, Richards H, Mullany P. Isolation of silver- and antibiotic-resistant Enterobacter cloacae from teeth. Oral Microbiol Immunol. 2005;20(3):191-194.
24. Silver S, Phung LT. Bacterial heavy metal resistance: new surprises. Annu Rev Microbiol. 1996;50:753-789.
25. Starodub ME, Trevors JT. Silver accumulation and resistance in Escherichia coli R1. J Inorg Biochem. 1990;39(4):317-325.
26. Gupta A, Matsui K, Lo JF, Silver S. Molecular basis for resistance to silver cations in Salmonella. Nat Med. 1999;5(2):183-188.
27. Silver S. Bacterial silver resistance: molecular biology and uses and misuses of silver compounds. FEMS Microbiol Rev. 2003;27(2-3):341-353.
28. Silver S, Gupta A, Matsui K, Lo J-F. Resistance to Ag(i) cations in bacteria: environments, genes and proteins. Metal-Based Drugs. 1999;6:315-320.
29. Silver S, Lo JF, Gupta A. Alliance for the Prudent Use of Antibiotics News. 1999;17(3):1-3.
30. Gupta A, Maynes M, Silver S. Effects of halides on plasmid-mediated silver resistance in Escherichia coli. Appl Environ Microbiol. 1998;64(12):5042-5045.
31. Silver S, Phung Le T, Silver G. Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds. J Ind Microbiol Biotechnol. 2006;33(7):627-634.
32. Newman GR, Walker M, Hobot JA, Bowler PG. Visualisation of bacterial sequestration and bactericidal activity within hydrating Hydrofiber wound dressings. Biomaterials. 2006;27(7):1129-1139.
33. Lansdown A, Williams A. Bacterial resistance to silver-based antibiotics. Nurs Times. 2007;103(9):48-49.
34. Bowler PG, Duerden BI, Armstrong DG. Wound microbiology and associated approaches to wound management. Clin Microbiol Rev. 2001;14(2):244-269.
35. Ip M, Lui SL, Chau SS, Lung I, Burd A. The prevalence of resistance to silver in a Burns unit. J Hosp Infect. 2006;63(3):342-344.













